|
Regulation
and Evolution of Transcription Initiation
The
DNA sequence of a gene needs to be copied into mRNA molecules to carry
out its function, a process called transcription. Cells are highly
selective in where they initiate the transcription of their genes. The
mechanism of locating the starting positions of transcription is shared
by most eukaryotic organisms, such as humans, plants, and fungi. Yet, a
distinct way of transcription initiation has been found in a few fungal
species including Baker's yeast. The main research focus of my lab is
to better understand the mechanisms of transcription initiation, and to
investigate how the conserved mechanisms have diverged in yeasts.
To acheive these goals, my lab generated a large
collection of high-throughput sequencing data to determine the
transcription start sites (TSS) at a single-nucleotide resolution for
different species and in response to environmental cues. In addition,
we built a user-friendly database
to visualize these TSS data, and
develop new software
for TSS data analysis.
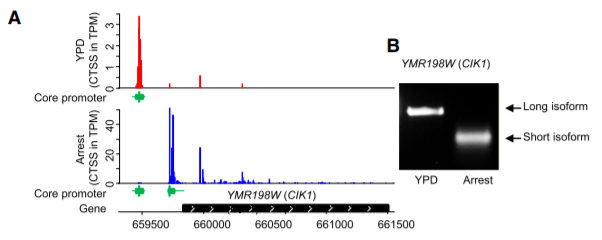
Most
eukaryotic genes use multiple clusters of TSS (core promoters). The use
of each core promoter is regulated in response to environmental cues
(called core
promoter shift).
(A) An example of core promoter shift (CIK1) between two growth
conditions, YPD and arrest. (B) Experimental validation displays the
presence and shift of two CIK1 transcript isoforms in response to
changing environments ( Lu
and Lin 2019 ).
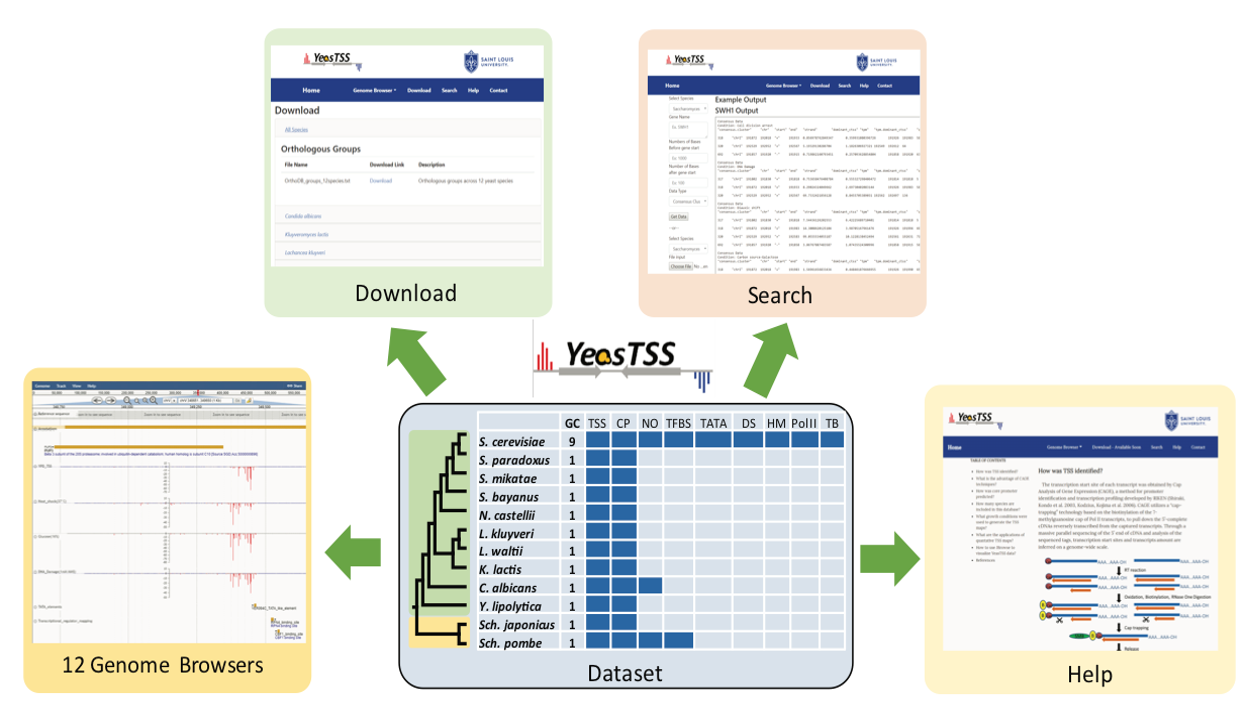
YeasTSS
is a public database that visulizes and integrates functional genomics
data relates to transcription regulation in different yeast species (Mcmillan
et al. 2019).
Evolution
of Genomes
An
organism’s genome contains the complete set of its genetic
information. Changes in the sequence and structure of a genome provide
raw genetic materials for functional innovation and evolutionary
divergence of living organisms. A long-standing question in
evolutionary biology is to determine the links between the genomic
variations and the evolution of new phenotypes of new species.
Another research focus of my lab is to better understand the
evolutionary patterns and mechanisms of the genomic sequences, gene
content and genome structures, and to learn their impacts on the
evolution of biological novelty and diversity through comparative
analyses of genome sequence data from different lineages of
organisms.
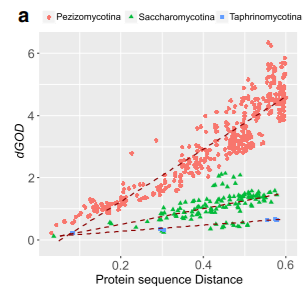
Our
study found that a higher rate of rearrangement at the genome scale
might have accelerated the speciation process and increased species
richness during the evolution ( Rajeh
et al. 2018. )
Ribosomal
protein (RP) genes encode structural components of ribosomes, the
cellular machinery for protein synthesis. We found that massive
duplications of RP genes have independently occurred by different
mechanisms. The sequences of most RP paralogs have been homogenized by
repeated gene conversion in each species, demonstrating parallel
concerted evolution, which might have facilitated the retention of
their duplicates (
Mullis
et al. 2018).
Our research program is supported by:
|





